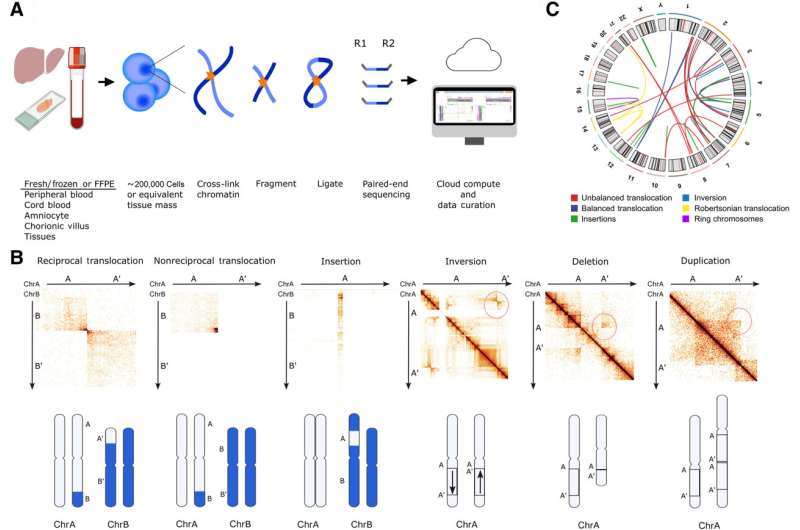
Traditional methods sequence DNA in a linear, one-dimensional way, reading the genetic code as if it were a flat line of text. In contrast, 3D chromosome mapping captures the spatial relationships between different parts of the genome. It reveals how the long strands of DNA fold and interact with each other in the three-dimensional space of the cell nucleus, which is vital for detecting certain structural changes that are invisible to conventional linear tests.
Researchers applied genomic proximity mapping (GPM), a genome-wide Hi-C (high-throughput chromosome conformation capture sequencing)-based NGS assay, to DNA from 123 individuals with suspected genetic disorders. This approach captured the 3D contacts in the genome, which allowed the detection of both copy-number changes and rearrangements in DNA. GPM correctly identified all known large chromosomal variants (110 deletions/duplications and 27 rearrangements) with 100% concordance. It also uncovered 12 novel structural variants that were missed by standard clinical tests.