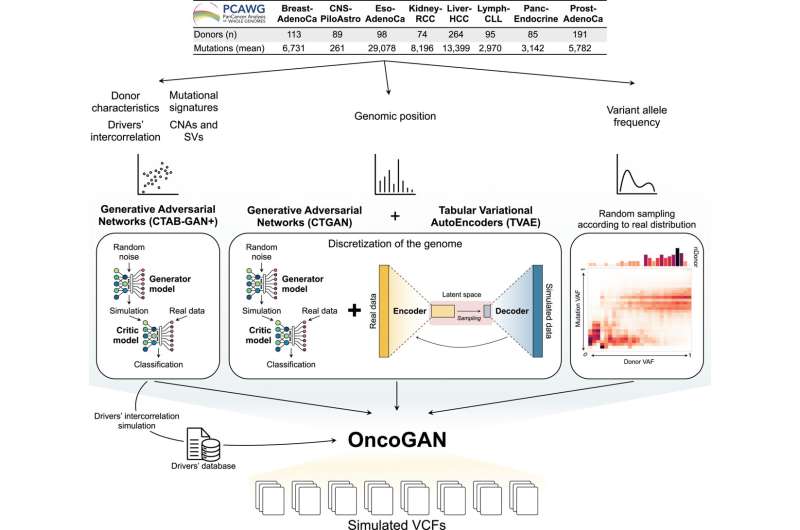
OncoGAN was developed by researchers at the Ontario Institute for Cancer Research (OICR) and the University of Toronto and is described in a new Cell Genomics paper.
It uses generative AI to simulate realistic tumor genomes across eight different types of cancer, including breast, prostate and pancreatic cancers. These synthetic genomes can simulate realistic patterns of genetic alterations, and can be used to benchmark genomic testing and improve the algorithms that make “precision oncology” possible.
Analyzing tumor genomes and the variations within their DNA has enabled new discoveries about how cancer develops, leading to a surge of cutting-edge tests and medicines. It is the cornerstone of precision oncology, where cancer treatment is personalized to the unique biology of a patient’s tumor.
But the algorithms used to analyze genomes are limited because they have been trained on a limited set of cancer genomes, relatively few of which are publicly available. The most commonly used tools were trained on a few dozen legacy genomes, and can’t fully capture the necessary biological diversity. While more recent genome sequencing data exists, access is often restricted due to concerns around the confidentiality of the patients they were sampled from.