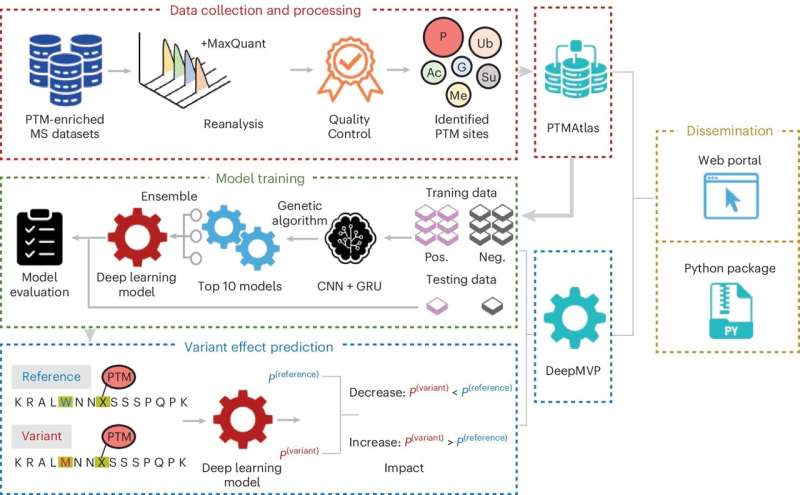
“Proteins are responsible for all the functions of the body, from growing tissues to regulating metabolism or fighting disease. Their functions are often regulated by modifications that take place after proteins are made through a process called post-translational modification (PTM),” said corresponding author Dr. Bing Zhang, professor at the Lester and Sue Smith Breast Center and of molecular and human genetics at Baylor. He also is a McNair scholar and a member of Baylor’s Dan L Duncan Comprehensive Cancer Center.
The modifications include the addition of chemical groups, such as phosphates or sugars, that influence how a protein behaves, where it goes in the cell or how long it lasts. When PTMs go wrong, the proteins may not perform as expected and contribute to diseases like cancer, heart conditions or neurological disorders.
Understanding where PTMs happen can help predict how mutations in these locations may change a protein’s function in ways that affect a person’s health. For instance, PTMs can be disrupted by DNA mutations that can remove a PTM site in a protein, create a new site or affect nearby regions, altering the protein’s function.